The Simulation tab allows the user to execute simulations for each of the scenarios specified for the study. The user may choose the number of simulations, whether to execute locally or on the Grid, and modify the random number seeds.
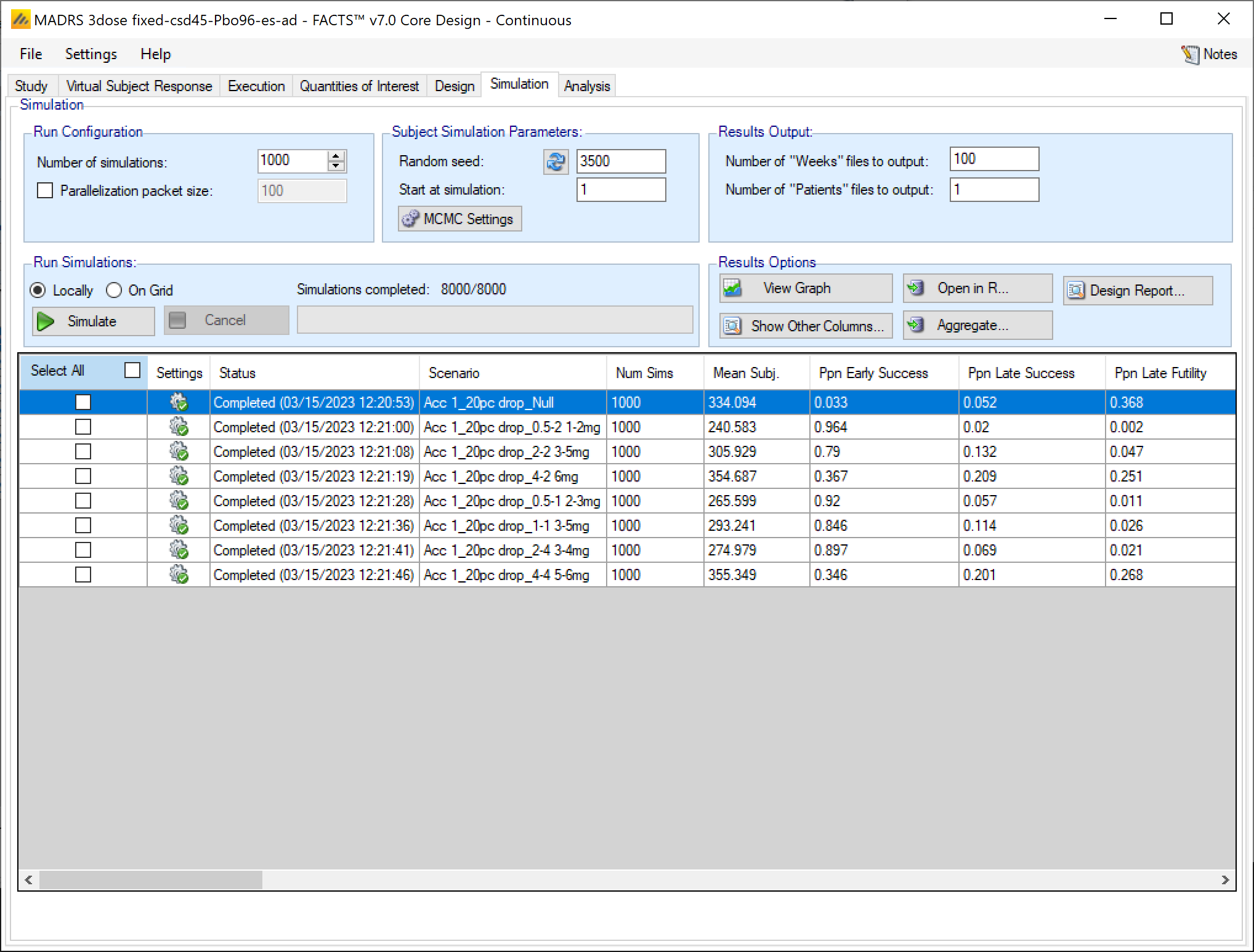
In the Simulation tab the user can provide simulation configuration parameters like the number of simulations to run, whether the simulations can be run on the Grid, the parallelization strategy, the random number seed used in the simulations, and the number of certain output files that should be kept during the simulation execution.
Simulation Options
Number of simulations
This box allows the user to enter the number of simulations that they would like FACTS to run for each scenario listed in the table at the bottom of the simulation tab. There is no set number of simulations that is always appropriate.
- 10 simulations
- You might want to run 10 simulations if you just want to look at a few simulated trials and assess how the decision rules work and if FACTS is simulating what you expected based on what you input on the previous tabs. If all 10 simulations of a ‘null’ scenario are successful, or all 10 simulations of what was intended to be an effective drug scenario are futile, it is likely there has been a mistake or misunderstanding in the specification of the scenarios or the final evaluation or early stopping criteria.
- 100 simulations
- You might want to run 100 simulations if you want to look at many individual trials to make sure that what you want to happen is nearly always happening. You can also start to get a very loose idea about operating characteristics like power based on 100 sims. 100 simulations is also usually sufficient to spot big problems with the data analysis such as poor model fits or significant bias in the posterior estimates.
- 1,000 simulations
- You might want to run 1,000 simulations if you want estimates of operating characteristics like power, sample size, and Type I error for internal use or while iterating the design. This generally isn’t considered enough simulations for something like a regulatory submission. With 1,000 simulations the standard error for a typical type I error calculation is on the order of \(0.005\).
- 10,000 simulations
- You might want to run 10,000 simulations per scenario if you are finalizing a design and are preparing a report. This is generally enough simulations for a regulatory submission, especially in non-null simulation scenarios. The standard error for a typical type I error calculation using 10,000 simulations is on the order of \(0.0015\).
- > 10,000
- You might want to run more than 10,000 simulations if you want to be very certain of an operating characteristic’s value - like Type I error. And plan to use the measurement of the quantity for something important like a regulatory submission. The standard error of a Type I error calculation with 100,000 simulations, e.g., is on the order of \(0.0005\).
- > 100,000
- You probably don’t want to run more than 100,000 simulations per scenario. Maybe your finger slipped an hit an extra 0, or you thought there were 5 zeroes in that number when there were actually 6. If the simulated trial is adaptive, this is going to take a while.
Each time the FACTS application opens, the “Number of Simulations” will be set to the number of simulations last run for this design. Not all scenarios must be run with the same number of simulations. If completed results are available, the actual number of simulations run for each scenario is reported in the ‘Num Sims’ column of the results table. The value displayed in the “Number of Simulations” control is the number of simulations that will be run if the user clicks on the ‘Simulate’ button.
Note also that if a scenario uses an external VSR file or directory of external files, the number of simulations will be rounded down to the nearest complete multiple of the number of VSR lines or external files. If the number of simulations requested is less than the number of VSR lines or external files, then just the requested number of simulations are run.
Start at Simulation
The “Start at simulation” option allows for the simulation of a particular trial seen in a previous set of simulations without having to simulate all of the previous trials in that previous set to get to it.
The initial random seed for FACTS simulations is set in the simulation tab. The first thing that FACTS does is to draw the random number seeds to use at the start of each simulation. Thus, it is possible to re-run a specific simulation out of a large set without re-running all of them. For example, say the 999th simulation out of a set displayed some unusual behavior, in order to understand why, one might want to see the individual interim analyses for that simulation (the “weeks” file), the sampled subject results for that simulation (the “Subjects” files) and possibly even the MCMC samples from the analyses in that simulation. You can save the .facts file with a slightly different name (to preserve the existing simulation results), then run 1 simulation of the specific scenario, specifying that the simulations start at simulation 999 and that at least 1 weeks file, 1 subjects file and the MCMC samples file (see the “MCMC settings” dialog) are output.
Parallelization Packet Size
The parallelization packet size option allows simulation jobs to be split into runs of no-more than the specified number of trials that are run in parallel. If more simulations of a scenario are requested than can be done in one packet, the simulations are broken into the requisite number of packets, run, and combined and summarized when they are all complete. The final results files will look just as though all the simulations were run as one job or packet.
The packet size must be a perfect divisor of the number of simulations. This is usually easy since common numbers of simulations are multiples of 100, but don’t try to use a prime number for Number of Simulations or you’re stuck with only 2 packet size options.
By default (if the check box with Choose Parallel Packet Size is not checked) the number of simulations per packet depends on the number of simulations per scenario. If the number of simulations is less than 1000, then each scenario is packaged as a single packed and simulated. If the number of simulations per scenario is greater than or equal to 1000, the default packet size is 10 and all simulations are decomposed into packets of size 10.
If an external file is used to create explicit VSRs (a .mvsr file), then the packet size should be a multiple of the number of rows in that MVSR file. Each packet will get passed the entire .mvsr file to run. If there are multiple .mvsr files with differing numbers of lines then only the VSR scenarios that have a .mvsr file that has a number of rows that is a divisor of the packet size will be run. The rest will error. The packet size can then be modified to get each of the .mvsr specified VSR files to be run.
Care should be taken when packetizing a scenario that includes an external data file to supply the virtual subject responses; in this situation, a of copy of the external file is included in each packet which can cause the packetisation process to run out of memory as the packets are being created. In this case, use a smaller number of larger packets, such as packets that are 1/10th of the total number of simulations.
When running simulations, FACTS will create and run as many packets in parallel as there are execution threads on the local machine. In general, the overhead of packetization is quite low, so a packet size of 10 to 100 can help speed up the overall simulation process. Threads used to simulate scenarios that finish quickly can pick up packets for scenarios that take longer. The progress bar updates as simulation packets complete, so the smaller the packet size, the more accurately FACTS can report the overall progress of the simulation execution.
Random Seed
Random number generation plays a huge role in FACTS’s virtual patient generation and statistical analyses. In order to exactly reproduce a statistical set of results, it is necessary to start the random number generation process from an identical “Random Seed”. Using the same random seed in the same version of FACTS guarantees that simulated trials will always be reproducible. Changing the design parameters or the version of FACTS may or may not remove this reproducibility depending on the change.
Even a small change in the random seed will produce very different simulation results.
In addition to setting the seed, the user can choose whether they want the “Same seed for all scenarios” or “Different seed” for different scenarios. If “Same seed for all scenarios” is selected, the subjects generated for each simulated trial will match for the different scenarios. This induces a correlation among the simulation output for different scenarios. This can be good if you’re trying to compare operating characteristics for different scenarios, but it can also be misleading. To disable this option select the “Different Seed” option. If “Different seed” is selected, then each scenario has its own seed that samples a different set of subjects than any other scenario. This uncorrelates the simulation output across scenarios, which can be advantageous if the absolute value of the operating characteristics are more valuable to you than the comparison of operating characteristics across scenarios.
MCMC Settings
To set advanced settings for simulation, the user may click the “MCMC Settings” button, which will display a number of additional specifiable parameters for simulation in a separate window.
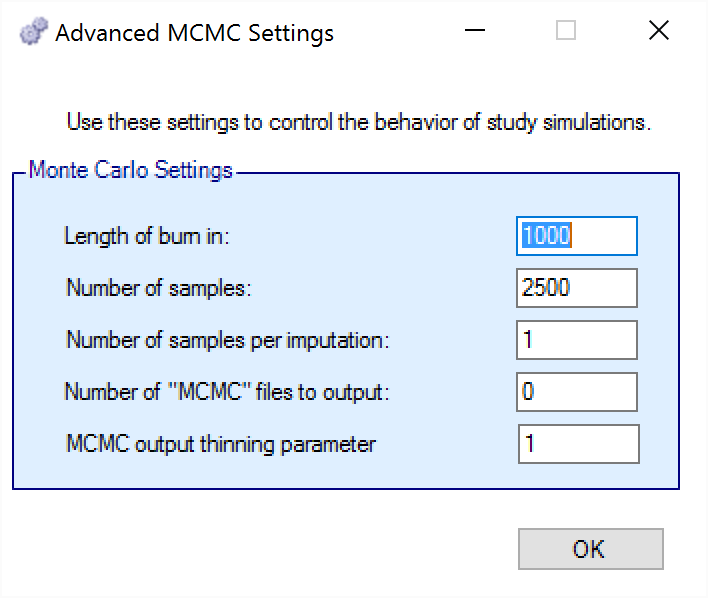
The first two values specify two standard MCMC parameters –
The length of burn-in is the number of the initial iterations whose results are discarded, to allow the MCMC chain to reach its stationary distribution. Burn-in samples are output in MCMC files if the files are output.
The number of samples is the number of subsequent iterations whose results are recorded in order to give posterior estimates of the values of interest.
The third parameter controls the number of MCMC samples taken after each imputation of missing data using the longitudinal model. The default value is 1. This parameter only has an effect if Bayesian imputation is being used to impute missing or partially observed data. Increasing the value of this parameter allows the parameter estimates to converge somewhat to a potentially new stationary distribution for each new set of imputed data. If the imputed data is only a small percentage of the overall data this is likely unnecessary. As a rough guide, if it at some early interims > 5% of the data being analyzed will be imputed, a value in the range 2 to 10 is recommended to avoid underestimating the uncertainty. A higher number should be used the greater the proportion of imputed data.
The next parameter concerns the output of the MCMC samples to a file. It is possible to have the design engine output the sampled values from the MCMC in all of the interims of the first N simulated trials of each scenario by specifying the “Number of MCMC files to output” to be greater than 0. The resulting files, ‘mcmcNNNN.csv’, will be in the results directory with all the other results files for that scenario. These files include the burn-in samples from the MCMC chains.
The final parameter in MCMC Settings is the thinning parameter. This parameter will only keep every \(N^{th}\) sample taken during MCMC where \(N\) is the thinning parameter. Thinning MCMC samples can reduce the autocorrelation of consecutive MCMC iterations, which increases the effective samples per retained sample, but also results in needing many more MCMC iterations to reach the same number of retained samples. Generally, we do not recommend thinning for standard simulation runs.
Unlike other software that performs MCMC, when you choose to thin by a value, FACTS does not increase the number of MCMC iterations it performs in order to retain the value specified in “Number of Samples”. So if you leave “Number of Samples” at its default value, \(2500\), and thin by \(10\), you will be left with \(250\) retained samples. You should adjust for this by increasing the “Number of Samples” if you choose to thin.
Results Output
The results output section of the Simulation tab allows for the specification of how many output files should be generated for files that are individually created for each simulation. Summary files (summary.csv) that have 1 line per scenario are always created. Simulations files (simulations.csv) that have 1 line per simulation are always created. Weeks files (weeksXXXXX.csv), patients files (patientsXXXXX.csv), and frequentist weeks files (weeks_freq_{missingness}_XXXXX.csv) are not created for every single simulation. Instead, the number of simulation specific output files can be set per type. This limits the amount of output files that FACTS will save.
See the endpoint specific descriptions of the output files for descriptions of what the previously mentioned output files report. See here for core continuous or dichotomous output files, and see here for core time-to-event output files.
Some plots in FACTS that are created based on weeks files, and if very few weeks files are saved, the plots will not be as accurate or descriptive.
Run Simulations
Click in the check box in each of the rows corresponding the to the scenarios to be run. FACTS displays a row for each possible combination of the ‘profiles’ that have been specified: - baseline response, dose response, longitudinal response, accrual rate, and dropout rate. Or simply click on “Select All”.
Then click on the “Simulate” button.
During simulation, the user is prevented from modifying any parameters on any other tab of the application. This safeguard ensures that the simulation results reflect the parameters specified in the user interface.
When simulations are started, FACTS saves all the study parameters, and when the simulations are complete all the simulation results are saved in results files in a “_results” folder in the same directory as the “.facts” file. Within the “_results” folder there will be a sub-folder that holds the results for each scenario.
FACTS Grid Simulation Settings
A user with access to a computational grid may choose to run simulations on the grid instead of running them locally. This frees the user’s computer from the computationally intensive task of simulating so that they can continue other work or even shutdown their PC or laptop. In order to run simulations on the grid, it must first be configured. This is normally done via a configuration file supplied with the FACTS installation by the IT group responsible for the FACTS installation.
Simulation Results
In the center of the simulation tab, the summary simulation results are displayed. There are many columns of results, these are organized into related groups of sub-windows, which can be displayed by clicking on the “Show More Columns” button.
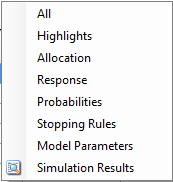
These windows will show:
| Name | Column Description |
|---|---|
| All | All summary columns |
| Highlights | Only the columns shown on the main tab |
| Allocation | The columns that report on participant recruitment and allocation |
| Response | The columns that report that estimate treatment response, the SD of the estimate, the estimate of the SD of the response, the true treatment response and the true SD of the response. |
| Observed | The raw endpoint output and the dropout rates by arm and visit |
| Probabilities | The final estimates for the QOIs that were computed for the trial. |
| Stopping Rules | The proportion of times the different stopping criteria were met |
| Model Parameters | The columns that report the estimates of the values of the model parameters. |
| Simulation Results | A window that displays the individual simulation results for the currently selected scenario. |
| Simulation Duration | A window that displays how long each scenario took to simulate |
| Frequentist results | If frequentist analysis is enabled, the summary results can be viewed, these are grouped by how missing data has been treated: Last Observation Carried Forward (LOCF), Baseline Observation Carried Forward (BOCF – if baseline has been simulated), Per-Protocol (PP). |
Open in R
The “Open in R” button allows for the creation of an R script that has pre-populated code for loading in output files created by the FACTS simulations.
By default, any/all of the simulation output files can be included in the created script. If “Aggregation” (see below) has been performed, then only the aggregated files will be available for being loaded in R.
When the button is clicked, FACTS will create an R script with the correct file paths to load in the data, as well as creating a function that will read the files in correctly. The file is then opened in the default R editor for the user. If there is no default program for opening a .R file, your operating system should ask how you want to open the file.
Aggregation
Aggregation combines the csv output from multiple scenarios into fewer csv files. The Aggregate… button displays a dialog which allows the user to select what to aggregate.
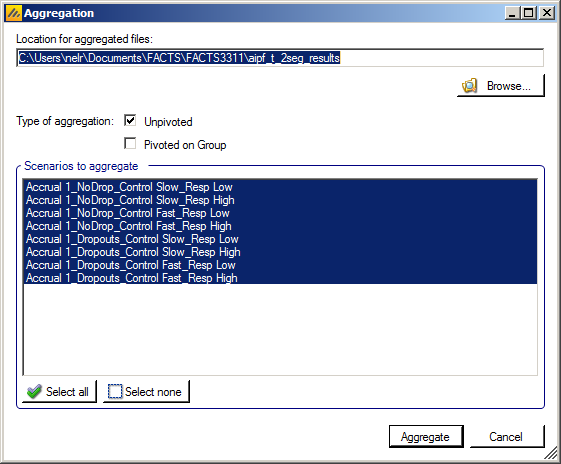
The default location for the aggregated files is the results directory for the study, but this can be changed.
Aggregation may be performed with or without pivoting on group, or both.
Unpivoted files will have one row for each row in the original files.
In pivoted files each original row will be split into one row per dose.
Where there is a group of columns for each dose, they will be turned into a single column with each value on a new row.
Values in columns that are independent of dose will be repeated on each row.
The default is to aggregate all scenarios, but any combination may be selected.
Pressing “Aggregate” generates the aggregated files.
Each type of csv file is aggregated into a separate csv file whose name begins agg_ or agg_pivot_, so agg_summary.csv will contain the rows from each of the summary.csv files, unpivoted. WeeksNNNNN.csv files are aggregated into a single agg_[pivot_]weeks.csv file. PatientsNNNNN.csv files are aggregated into a single agg_patients.csv file, but they are never pivoted because each row already refers to a single dose. Similarly the various frequentist results at the summary, simulation and weeks level are aggregated (if they’ve been output).
RegionIndex.csv is not aggregated.
Each aggregated file begins with the following extra columns, followed by the columns from the original csv file:
| Column Name | Comments |
|---|---|
| Scenario ID | Index of the scenario |
| Recruitment Profile | A series of columns containing the names of the various profiles used to construct the scenario. Columns that are never used are omitted (e.g. External Subjects Profile if there are no external scenarios) |
| Dropouts Profile | |
| Longitudinal Rates Profile | |
| Dose Response Profile | |
| External Subjects Profile | |
| Agg Timestamp | Date and time when aggregation was performed |
| P(TS) | Proportion of trial success (early success + late success) |
| P(TF) | Proportion of trial futility (early futility + late futility) |
| Sim | Simulation number. Only present in weeks and patients files. |
| Dose | Only present if pivoted |
Design Report
This button becomes enabled once there are simulation results, it uses an R script and R libraries to generate a MS Word document describing the design.
See the FACTS Design Report User Guide for details of what R packages need installing, how FACTS needs configuring to use the correct R instance, how the generate_report() function is run, and where the resulting report can be found.
